Classification of AML and CML
The French-American-British (FAB) classification for AML was based on cytomorphologic features and has been replaced by the WHO 2001/2008 and 2016 classifications.
The European LeukemiaNet (ELN) defines 3 risk groups according to genetic abnormalities (Döhner et al. 2017).
Certain AML subgroups such as acute promyelocytic leukaemia (APL) (PML-RARA, t[15;17]) benefit from targeted treatment and have an excellent prognosis.
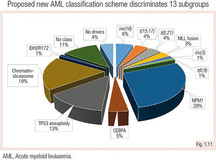
A new classification scheme was proposed, including karyotype and somatic mutations, and defines 13 AML subgroups (Papaemmanuil et al. 2016).
Specific chromosomal aberrations such as t(8;21), inv(16), t(15;17) are disease-defining, irrespective of the quantified blast count.
In the future, diagnosis of AML might rely solely on genetic findings.
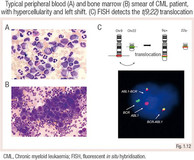
CML is characterised by leukocytosis with myeloid progenitors in the peripheral blood termed ‘left shift’. The disease is driven by the Philadelphia chromosome t(9;22), which produces the constitutive active fusion protein BCR-ABL1.
CML is classified into chronic phase, accelerated phase and blast phase, according to the blast cell count.
Therapy monitoring is performed using highly sensitive real-time PCR to detect BCR-ABL1.
Revision Questions
- What is the basis for the WHO 2016 AML classification?
- Which aberrations and mutations are diagnostic for AML without a need for ≥20% blasts?
- What is the genetic basis of CML?