Microsatellite instability (MSI) indicates a defective mismatch repair (dMMR) system
DNA mismatch repair
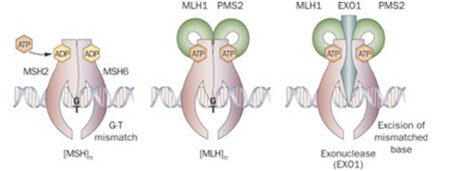
DNA mismatch repair is a highly conserved mechanism, involved in restoring DNA integrity after the occurrence of mismatching errors during DNA replication, recombination or iatrogenic damage1,2,3. Four genes regulate the MMR mechanism: mutL homologue 1 (MLH1), mutS homologue 2 (MSH2), mutS homologue 6 (MSH6) and postmeiotic segregation increased 2 (PMS2). The four proteins codified by these genes from heterodimers, namely MLH1/PMS2 and MSH2/MSH6 (see image below). The biallelic inactivation of one of these genes, due to somatic or germline mutations or epigenetic silencing, results in dMMR determining an increased rate of mutations1,4,5.
Microsatellites
Microsatellites, or short tandem repeats, are repetitive DNA sequences with a unit length ranging from one (mononucleotides) to six bases (di-, tri-, tetra-, penta-, esa-nucleotides) distributed along coding and noncoding regions of the genome4. They are highly polymorphic among subjects but stable in each individual6,7. The repetitive nature of these regions makes them particularly sensitive to mismatch errors that in case of dMMR result in the accumulation of mutations of repeat length alterations, defined as microsatellite instability (MSI), which is easily uncovered by the analysis of polyA microsatellites. Therefore, MSI is a marker of dMMR6,7 and characterizes a hypermutable state of cells8.
Definition of MSI/dMMR tumour
A tumour with MSI has thousands of mutations due to a defective DNA mismatch repair (dMMR) system, caused by biallelic inactivation of one of the four genes coding for the proteins involved in this mechanism. MSI is efficiently detected by a molecular test analysing few polyA DNA microsatellites that, due to their monomorphic composition, are highly prone to misalignments during DNA replication.
Clinical significance of MSI-dMMR
MSI testing assesses the functionality of the MMR system and has different clinical significance for sporadic and hereditary cancers. It has an established role in the identification of hereditary cancer syndromes and is of prognostic significance in surgically resected gastrointestinal cancers. It also has an emerging potential predictive value of response to immunotherapy 10,11,12. These findings have recently increased the clinical request for MSI molecular testing as a predictive biomarker for immunotherapy, increasing the need of a more targeted assessment of truly necessary exams to avoid excessive and fruitless costs for the health system.
MSI testing to detect dMMR
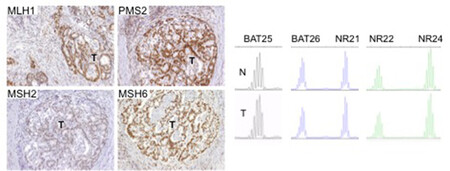
There are two clinically useful tests to detect a dMMR in cancer (see images below): i) identification of MSI by molecular testing of poly-A microsatellites, which represents direct proof of dMMR; ii) lack of immunohistochemical expression of MMR proteins that represents an indirect suggestion of a dMMR system, which should be confirmed with MSI molecular testing.
MSI molecular test
Molecular testing is performed on DNA from fresh, frozen, or paraffin-embedded tumour tissue using a PCR-based assay for detection of MSI. The highest specificity and sensitivity is reached using a panel of three or more polyA mononucleotide markers (BAT25, BAT26, NR-21, NR-22, NR-24, NR-27) 4,13,14.
Immunohistochemistry (IHC)
Antibodies against the four MMR proteins are commercially available and IHC is used to detect the expression of the four MMR proteins (MLH1, MSH2, MSH6, and PMS2), whose loss is highly concordant (>90%) with DNA-based MSI positive testing with good specificity and sensitivity4,15,16,17. As said above, the 4 proteins involved form MLH1/PMS2 and MSH2/MSH6 heterodimers (Image Model of the Proposed Mechanism of Mismatch Repair Proteins). The loss of expression of a single protein or of a heterodimer couple suggests the presence of dMMR, which requires formal proof by MSI molecular testing, as IHC loss of expression may be a false result due to technical or biological reasons. Moreover, an inhomogeneous staining may occur for one or more of the four antibodies again to technical or biological reasons18.In cancers with dMMR proved by molecular testing, IHC loss of expression is used to indicate which gene is defective due to germline or somatic mutation or inactivation by hypermethylation19. The major advantage of IHC is its wide and routine availability in pathology laboratories. Limitations of IHC include misleading information for rare cases with missense mutations in MLH1 or MSH6 genes with a normally translated but non-functional protein. In these cases positive IHC implies a false negative result for dMMR, and only molecular MSI testing can clarify the real functionality of MMR19. On the other hand, IHC detects MSH6 loss in a subset of MSH6 germline mutated tumours, implying a false positive result for dMMR where, however, the presence of a functional redundancy in the MMR system does not affect the proficiency of the MMR machinery, and this can only be clarified by a negative MSI molecular test19. Technical reasons may also cause false negative IHC results.
Choice of test
Molecular testing of polyA microsatellites is the choice to detect MSI as direct proof of dMMR in a given cancer. Immunohistochemistry can be used as an efficient indirect test for dMMR when a molecular laboratory is not available; it is also useful in identifying which gene should be investigated for damaging alterations in the case of hereditary cancer syndromes15.
Confounding definitions of MSI in the literature of the last two decades
There is much confusion in the literature of the last couple of decades due to the varying definitions of MSI used, linked to the plethora of different mononucleotide and polynucleotide microsatellite markers used by the diverse authors and in different cancer types.
In 1993 Manuel Perucho’s team20 reported the discovery of colorectal cancers harbouring ubiquitous somatic mutations (over 100,000) in simple repeated sequences, due to mutations in a DNA replication factor resulting in reduced fidelity for replication or repair. It was clearly reported that only tumours with affected polyA microsatellites carry mutations in other polynucleotide simple repeats20. Five years later, a consensus workshop at Bethesda recommended a “NCI-reference panel” to assess MSI including two mononucleotides (BAT25 and BAT26) and three dinucleotides (D2S123, D5S346, D17S250), whose status in cancer DNA had to be compared with DNA from normal tissue of each patient21. On the basis of the results of such a molecular test, tumours were classified into three different subtypes: MSI-high (MSI-H), if two or more microsatellite markers showed instability; MSI-low (MSI-L) if only one marker resulted unstable and MS-stable (MSS) if all the five markers resulted stable21. The revision of these guidelines based on evidence that only MSI-H should be considered for the definition of true MSI tumours, and only if two positive markers were polyA microsatellites, suggested the use of a panel including three or more polyA markers22. In fact, alterations in non-monotypic microsatellites (from di- to esa-nucleotides) may relate more to generalized chromosomal instability than to a deficient mismatch repair system23. The contrasting evidence around clinicopathological differences between MSI-L and MSS is likely to be attributed to the use of variable MSI markers and definitions. Thus it is often impossible to distinguish between MSI-L and chromosomal instability, suggesting that these tumours be classified as a single molecular subset, i.e. MSS neoplasms 4,22,24,25,26,27.
Hereditary cancer syndromes associated with MSI cancers
Lynch syndrome. Hereditary Non-Polyposis Colorectal Cancer (HNPCC) or Lynch syndrome (LS) is the most frequent inherited cancer predisposition syndrome caused by a germline heterozygous mutation in one of the four MMR genes28. These patients are characterized by early onset of tumours (average age <45 years), mainly colorectal and endometrial but also tumours in other organs, and usually present germline mutations in MLH1 or MSH2 (42% and 33%, respectively), followed by MSH6 and PMS2 (18% and 7%, respectively)29. Several guidelines have been developed to identify HNPCC patients, from the Amsterdam guidelines30 through the Bethesda guidelines22, to the Jerusalem Criteria31, together with the suggestion to implement a universal testing of MSI in colorectal cancers19,32,33.
Lynch syndrome due to TACSTD1 germline mutations. Another molecular alteration identified in HNPCC families is the heritable epigenetic silencing of MSH2 due to heterozygous germline deletion of exon 3 of TACSTD1 gene encoding for epithelial cell adhesion molecule (EpCAM)34.
Biallelic mismatch repair deficiency syndrome. Inherited homozygous mutations in any of the MMR genes identify this clinical syndrome characterized by gastrointestinal and brain tumours and haematologic malignancies along with café-au-lait macules in childhood and adults35.
Muir-Torre syndrome. Germline mutations in MSH2 and MLH1 genes have also been identified in families with Muir-Torre syndrome characterised by sebaceous gland tumours together with internal malignancies, commonly colorectal36.
Turcot's syndrome is clinically characterized by the early occurrence of primary brain and colorectal tumours and is caused by germline mutations in APC, MLH1 or PMS2 genes36.
MSI in different sporadic cancer types
Unfortunately, the variable definition of MSI and the plethora of markers and cut-off values used in the literature of the past 20 years have largely misguided the interpretation of published results, especially for non-colorectal tumours. In this factsheet, we revised the most pertinent literature on diverse cancers trying to discern the identification of true dMMR tumours, i.e. only those showing MSI-H and/or alterations in mononucleotide microsatellite markers.
Sporadic gastrointestinal cancers
Colorectal Cancer (CRC). About 15% of sporadic CRCs harbour MSI, the vast majority of which is related to MLH1 promoter hypermethylation, with subsequent silencing of gene transcription and loss of protein expression 5,15,37,38,39. The Association for Molecular Pathology recommends to subject all new colorectal cancers to MSI analysis to classify them into three subgroups: sporadic MMR-proficient, sporadic dMMR, or Lynch dMMR15. MSI sporadic CRCs are characterized by specific clinicopathological features: mainly female gender, older age, right colon location, high grade, mucinous differentiation, signet ring or medullary histology, peritumoural lymphocytic infiltrate and Crohn-like inflammatory reaction, diploid status, lower stage and better prognosis40. MSI is considered a favourable prognostic factor in early stage CRCs, with longer disease free and overall survival (DFS and OS) 41,42,43,44,45. Some authors hypothesize that their better prognosis may be partly explained by the increased immune response found in dMMR neoplasms45. In the subset of metastatic CRC (mCRC) dMMR has a lower prevalence (5%) compared to early CRCs and is associated with a poor prognosis, possibly due to the higher incidence of BRAFV600E mutation in comparison to proficient MMR mCRC46. Preclinical and clinical studies have demonstrated that dMMR negatively affects the response of CRCs to chemotherapeutics such as pyrimidine analogues, cisplatin, temozolomide and procarbazine. Moreover, fluorouracil-based adjuvant chemotherapy seems to improve patient outcome, in particular those with stage II colon cancer with MSS/MSI-L tumours but not those with tumours exhibiting MSI-H41,42,47. A retrospective analysis has shown a statistically significant survival benefit for patients with dMMR tumours by the addition of bevacizumab to adjuvant FOLFOX therapy compared with patients with proficient MMR tumours48. This data, deriving from a subgroup analysis, are preliminary and further investigation is needed particularly in metastatic setting. The utility of MSI status as a promising predictive marker for response to anti-PD-1 therapy in stage IV CRCs has been recently reported10,11. Furthermore, a high frequency of Th1/CTL (cytotoxic T lymphocyte) infiltrate (TILs) has been detected in MSI CRCs associated with up-regulation of at least five immune checkpoint molecules, targets of inhibitors whose efficacy is under clinical testing49. MSI can be acquired during chemotherapy by selective mutations in MMR genes2,3.
Gastric cancer (GC). GC shows MSI in about 15% of cases, which is usually associated with female sex, older age, antral location, intestinal histology, earlier stage and better prognosis14,50,51,52. The incidence of gastric cancer in HNPCC is low53. Although some conflicting results exist 4, MSI in gastric cancer may be considered a favourable prognostic indicator4,51,54 for both early14,52,55,56 and advanced stages57,58. A study including 12 MSI and 64 MSS surgically resected GCs treated with adjuvant 5-fluorouracil (5-FU) therapy reported no difference between MSI and MSS with respect to OS59. However, a more recent study including 796 (61 MSI and 735 MSS) resected GC patients reported at multivariate analysis that MSS patients had benefit from 5FU-based adjuvant chemotherapy in term of DFS and proposed MSI status as a predictive biomarker for 5-FU-based adjuvant chemotherapy in stage II and III gastric cancers after R0 resection60. Recently significant correlations have been found between dMMR and immune system activity, suggesting that this group of patients might be optimal candidates for novel immunotherapies61,62,63.
Duodenal and Ampulla of Vater Cancer (AVC). Duodenal cancers and AVCs show MSI in up to 10% of cases64,65,66,67,68,69. In AVC, MSI is associated with intestinal mucinous subtype, high-grade and markedly increased TILs65,66, even if a noticeable subset of MSS AVCs with higher TIL counts occurs66.
Esophageal carcinoma. MSI is restricted to Barrett’s-related esophageal adenocarcinomas, accounting for about 5% of this cancer type. It is associated with increased TILs and specific histotypes (medullary, signet ring cell, mucinous histology) similar to MSI colorectal cancers70,71, and has been suggested as potential candidate to immunotherapy62.
Sporadic gynaecological cancers
Endometrial cancer (EC). EC is associated with defective MMR in up to 33% of cases4,10, a proportion of which is related to HNPCC syndrome. Morphological heterogeneity, non-endometrioid histology and marked inflammatory response are distinctive characters of MSI-ECs72. The universal screening of ECs for MSI has been suggested to identify Lynch syndrome patients73,74. There is controversial evidence on the prognostic value of MSI in EC4,75,76,77. A pooled analysis of 22 studies failed to identify a significant association between prognosis and MMR status, although the marked inter-study heterogeneity regarding patients’ characteristics, tests used for detecting MSI, and estimation of endpoints make it difficult to establish an exact role of dMMR in EC78. MSI-ECs showed a high rate of PD-1/PD-L1 overexpression plus enhanced TILs, suggesting that MSI-ECs might represent the perfect candidates for PD-1 directed immunotherapies79.
Ovarian cancer (OC). When considering only tumours tested for MSI using panels containing polyA mononucleotides the value of MSI in OCs is 10% 4,80,81,82. Germline mutations in MMR genes are detected in about 1% of cases4,82,83,84. OC arising in women ≤ 50 years showed MSI in about 4% of cases, suggesting that these should be tested for MSI and, if unstable, for germline mutations in MMR genes80. Inconsistent data have been reported regarding both the prognostic significance 4,85,86,87 and predictive value 4,88,89 of MSI.
Cervical Cancer. MSI occurs in about 5% of cervical cancers90,91 of any morphology, but possibly in higher proportions in squamous cell (SCC) histotype (11.8%)92. No prognostic or predictive value seems to be associated with MSI status in this pathology90,92.
Breast cancer. MSI is extremely rare in breast cancer ranging between 0-1% of cases10,93, unless occurring in young women diagnosed with HNPCC94,95.
Sporadic hepatic, pancreatic and biliary tract cancers
MSI is exceedingly rare if not absent at all in hepatic, biliary tract and pancreatic cancers.
Hepatocellular carcinoma. Only MSI-L has been reported in hepatocellular carcinoma96 and alterations in MMR genes are not implicated in its pathogenesis97.
Pancreatic and periampullary cancers, including terminal bile duct cancers. MSI is almost non-existent in sporadic pancreatic ductal adenocarcinoma (PDAC) occurring in less than 1% of cases based on molecular MSI testing. A study on 338 sporadic pancreatic ductal adenocarcinomas using mononucleotide repeats showed MSI in only 1 case, which involved the pancreatic head, ampulla of Vater and duodenum98. A recent study of 385 pancreatic cancers subjected to whole genome or exome sequencing reported 4 cases with MSI99. MSI is also found in the peculiar and rare medullary subtype of pancreatic carcinoma, where it was found in 4 of 18 (22%) tumours 100. The rare pancreatic cancers occurring as part of HNPCC show MSI. A recent paper reported that 15% of pancreatic cancers showed loss of MMR proteins using immunohistochemistry but no proof of defective MMR machinery in these cases was furnished101. A paper reporting MSI in 17% of cases used no polyA microsatellites markers102. While a paper reporting MSI in 13% of 100 cases using polyA markers which were also associated with a better prognosis, there was no distinction made regarding the site of origin of the cases and it is highly likely that the cases in this series were ampullary cancers103, as also suggested by two recent studies of exome sequencing on periampullary cancers originating from ampulla of Vater, terminal bile duct or terminal pancreatic duct where up to 10% of cases had MSI68,69.
Sporadic skin tumours and melanoma
Sebaceous skin tumour. Sebaceous gland skin tumours (sebaceous hyperplasias, sebaceous adenomas, and sebaceous carcinomas) are ‘sentinel’ pathologies of Muir-Torre syndrome. About 25% of sporadic sebaceous skin tumours show MSI 104,105. Due to such high prevalence MSI testing is recommended in all sebaceous neoplasms regardless of patient's age or other clinical characteristics106.
Melanoma. The use of mononucleotide markers and properly defined MSI status in melanoma has been largely neglected, and explains the reported highly variable prevalence of MSI, ranging from 2% to 30% of primary cases and up to 77% in metastatic tumours4,107. As such, also the information that MSI status increases from benign nevi (0%) through primary melanoma (11%) to metastatic melanoma (21%-77%) is largely unreliable and cast doubts on its utility to identify patients candidate to immunotherapy 4,107,108,109,110. The data for this neoplasms are too scant and the techniques used to define MSI are variable and questionable to draw definitive conclusions.
Other sporadic cancers
Lung cancer. Discrepancy in reported MSI prevalence in non-small cell lung cancer (NSCLC), ranging from 0% to 40%, is due to the different methodologies and cut-off values used to define MSI111,112,113. When consented mononucleotide markers are used, MSI is absent in SCLC (0%) and exceedingly rare in NSCLC (0-1%), and thus no prognostic or predictive value of dMMR status exist in lung cancer114,115,116.
Glioma. MSI is extremely rare (0.16%) in gliomas of adults117,118. Controversial data exist on MSI in gliomas of paediatric age, adolescents and young adults, although all studies were performed using mononucleotide markers and a proper definition of MSI117,118,119,120,121. MSI was reported in a significant proportion (between 18% and 33%) of high grade, paediatric and young adult gliomas, also in the setting of Turcot’s syndrome119,120 while other authors reported absence or a very low rate of MSI in paediatric and young adult gliomas (low or high grade)117,121. In these papers, the syndromic or sporadic nature of the tumours analysed is not always clearly addressed.
Prostate cancer. In prostate cancer, MSI using mononucleotide markers and somatic mutations involving MMR genes has been reported in a subset of tumours ranging from 1% in primary to 12% in metastatic cancers10,122,123. Males with HNPCC have a fivefold increased risk of developing prostate cancer124.
Thyroid cancer. MSI due to dMMR does not have a role in thyroid cancer pathogenesis. The highly discordant data about MSI, ranging from complete absence125,126 to 63% of cases, again depends on varying markers and definitions used in the different papers127,128,129.
Head and neck squamous cell cancer. In only around 1% of samples there is evidence of true MSI revealed when considering consented mononucleotide markers 130, while largely variable MSI definitions and markers used are responsible for the extreme range of frequencies (from 3% to 88%)130,131.
Renal cell carcinoma. MSI is practically absent in renal cell carcinoma (0%-0.7%)132,133.
Sarcoma. Reported MSI in soft tissue sarcoma and Ewing sarcoma appear to relate more to generalized genomic instability rather than to a dMMR system, and true MSI phenotype is not detected in sarcomas23,134,135.
MSI and immunotherapy
Use of immune checkpoint blockade molecules is a promising treatment even in advanced cancers resistant to all other chemotherapeutics10,136,137,138. It has recently been demonstrated that tumours with high TILs usually harbour dMMR resulting in MSI with a significant upregulation of immune checkpoint proteins10,49, suggesting that an increased mutational burden in MSI tumours leads to the creation of neoepitopes responsible for the immune response10,49. Furthermore, the effectiveness of MSI status as a predictive marker for response to PD-1 blockade was recently reported in stage IV colorectal cancer patients10,11.
Indeed, somatic hypermutation creating putative neoepitopes is generated not only by MSI/dMMR but also by a high mutational load of nonsynonymous mutations139,140,141 due to mutations in DNA polymerases POLE or POLD1, exposure to exogenous (cigarette smoke, UV radiation) and endogenous mutagens10,142,143. Such hypermutation may reveal higher predictive power than MSI status as immunotherapy response biomarkers. However, they rely to date on whole-exome sequencing and mass spectrometry of tumour samples followed by extensive bioinformatic analysis, an approach not yet practical for routine diagnostics10. By contrast, MSI molecular testing is already largely diffuse at the clinical level, which increases its potential as a ready-to-use approach to predict immunotherapeutic response in patients who have failed conventional therapy10.
Conclusions:
- Microsatellite instability is the effect of a defective mismatch repair machinery. Thus, the molecular test for MSI is the gold standard diagnostic tool to directly assess the proficiency of mismatch repair system.
- Lack of immunohistochemical expression of one or more of the four proteins involved in MMR (MLH1, MSH2, MLH6, PMS2) is an indirect indication of a defective MMR which needs confirmation with a molecular test for MSI.
- The MSI molecular test must be based on mononucleotide (polyA) markers. The use of polynucleotide markers should be discontinued as their alteration is often due to chromosomal instability rather than to mismatch repair alterations.
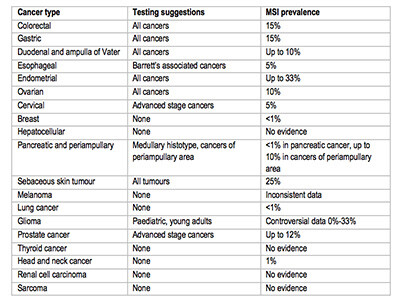
- MSI testing suggestions based on available data are reported in the image below.
References
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem 1996; 65: 101-133.
- Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res 2008; 18: 85-98.
- Kawakami H, Zaanan A, Sinicrope FA. Implications of mismatch repair-deficient status on management of early stage colorectal cancer. J Gastrointest Oncol 2015; 6: 676-684.
- Richman S. Deficient mismatch repair: Read all about it (Review). Int J Oncol 2015; 47: 1189-1202.
- Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A 1998; 95: 6870-6875.
- Arzimanoglou, II, Gilbert F, Barber HR. Microsatellite instability in human solid tumors. Cancer 1998; 82: 1808-1820.
- Weissenbach J, Gyapay G, Dib C, et al. A second-generation linkage map of the human genome. Nature 1992; 359: 794-801.
- Gatalica Z, Vranic S, Xiu J, et al. High microsatellite instability (MSI-H) colorectal carcinoma: a brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer 2016; 15: 405-412.
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Onc 2010; 7: 153-162.
- Dudley JC, Lin MT, Le DT, et al. Microsatellite Instability as a Biomarker for PD-1 Blockade. Clin Cancer Res 2016; 22: 813-820.
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015; 372: 2509-2520.
- Kelderman S, Schumacher TN, Kvistborg P. Mismatch Repair-Deficient Cancers Are Targets for Anti-PD-1 Therapy. Cancer Cell 2015; 28: 11-13.
- Buhard O, Cattaneo F, Wong YF, et al. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol 2006; 24: 241-251.
- Beghelli S, de Manzoni G, Barbi S, et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery 2006; 139: 347-356.
- Funkhouser WK, Jr., Lubin IM, Monzon FA, et al. Relevance, pathogenesis, and testing algorithm for mismatch repair-defective colorectal carcinomas: a report of the association for molecular pathology. J Mol Diagn 2012; 14: 91-103.
- Lindor NM, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002; 20: 1043-1048.
- Mills AM, Longacre TA. Lynch Syndrome Screening in the Gynecologic Tract: Current State of the Art. Am J Surg Pathol 2016; 40: e35-44.
- Shia J, Klimstra DS, Nafa K, et al. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colocrectal neoplasms. Am J Surg Pathol 2005; 29: 96-104.
- Zhang X, Li J. Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol 2013; 5: 12-19.
- Ionov Y, Peinado MA, Malkhosyan S, et al. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993; 363: 558-561.
- Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998; 58: 5248-5257.
- Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 2004; 96: 261-268.
- Monument MJ, Lessnick SL, Schiffman JD, et al. Microsatellite instability in sarcoma: fact or fiction? ISRN Oncol 2012; 2012: 473146.
- Jass JR, Young J, Leggett BA. Biological significance of microsatellite instability-low (MSI-L) status in colorectal tumors. Am J Pathol 2001; 158: 779-781.
- Tomlinson I, Halford S, Aaltonen L, et al. Does MSI-low exist? J Pathol 2002; 197: 6-13.
- Lee SY, Kim DW, Lee HS, et al. Low-Level Microsatellite Instability as a Potential Prognostic Factor in Sporadic Colorectal Cancer. Medicine (Baltimore) 2015; 94: e2260.
- Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol 2015; 16: 30.
- Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis 2008; 29: 673-680.
- Plazzer JP, Sijmons RH, Woods MO, et al. The InSiGHT database: utilizing 100 years of insights into Lynch syndrome. Fam Cancer 2013; 12: 175-180.
- Vasen HF, Watson P, Mecklin JP, et al. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 1999; 116: 1453-1456.
- Boland CR, Shike M. Report from the Jerusalem workshop on Lynch syndrome-hereditary nonpolyposis colorectal cancer. Gastroenterology 2010; 138: 2197 e2191-2197.
- Ladabaum U, Wang G, Terdiman J, et al. Strategies to identify the Lynch syndrome among patients with colorectal cancer: a cost-effectiveness analysis. Ann Intern Med 2011; 155: 69-79.
- Lynch PM. The Case for Universal Testing of Colorectal Tumors for Microsatellite Instability: A Coming Mismatch Between Clinical and Laboratory Testing. Dig Dis Sci 2015; 60: 2225-2227.
- Ligtenberg MJ, Kuiper RP, Chan TL, et al. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3' exons of TACSTD1. Nat Genet 2009; 41: 112-117.
- Durno CA, Sherman PM, Aronson M, et al. Phenotypic and genotypic characterisation of biallelic mismatch repair deficiency (BMMR-D) syndrome. Eur J Cancer 2015; 51: 977-983.
- Lawes DA, SenGupta S, Boulos PB. The clinical importance and prognostic implications of microsatellite instability in sporadic cancer. Eur J Surg Oncol 2003; 29: 201-212.
- Boissiere-Michot F, Frugier H, Ho-Pun-Cheung A, et al. Immunohistochemical staining for p16 and BRAFV600E is useful to distinguish between sporadic and hereditary (Lynch syndrome-related) microsatellite instable colorectal carcinomas. Virchows Arch 2016; 469: 135-144.
- Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 1997; 57: 808-811.
- Poynter JN, Siegmund KD, Weisenberger DJ, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev 2008; 17: 3208-3215.
- Setaffy L, Langner C. Microsatellite instability in colorectal cancer: clinicopathological significance. Pol J Pathol 2015; 66: 203-218.
- Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 2003; 349: 247-257.
- Guastadisegni C, Colafranceschi M, Ottini L, et al. Microsatellite instability as a marker of prognosis and response to therapy: a meta-analysis of colorectal cancer survival data. Eur J Cancer 2010; 46: 2788-2798.
- Malesci A, Laghi L, Bianchi P, et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res 2007; 13: 3831-3839.
- Muller CI, Schulmann K, Reinacher-Schick A, et al. Predictive and prognostic value of microsatellite instability in patients with advanced colorectal cancer treated with a fluoropyrimidine and oxaliplatin containing first-line chemotherapy. A report of the AIO Colorectal Study Group. Int J Colorectal Dis 2008; 23: 1033-1039.
- Tikidzhieva A, Benner A, Michel S, et al. Microsatellite instability and Beta2-Microglobulin mutations as prognostic markers in colon cancer: results of the FOGT-4 trial. Br J Cancer 2012; 106: 1239-1245.
- Venderbosch S, Nagtegaal ID, Maughan TS, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res 2014; 20: 5322-5330.
- Sargent DJ, Marsoni S, Monges G, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol 2010; 28: 3219-3226.
- Pogue-Geile K, Yothers G, Taniyama Y, et al. Defective mismatch repair and benefit from bevacizumab for colon cancer: findings from NSABP C-08. J Natl Cancer Inst 2013; 105: 989-992.
- Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov 2015; 5: 43-51.
- Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014; 513: 202-209.
- Falchetti M, Saieva C, Lupi R, et al. Gastric cancer with high-level microsatellite instability: target gene mutations, clinicopathologic features, and long-term survival. Hum Pathol 2008; 39: 925-932.
- Fang WL, Chang SC, Lan YT, et al. Microsatellite instability is associated with a better prognosis for gastric cancer patients after curative surgery. World J Surg 2012; 36: 2131-2138.
- Engel C, Loeffler M, Steinke V, et al. Risks of less common cancers in proven mutation carriers with lynch syndrome. J Clin Oncol 2012; 30: 4409-4415.
- Marrelli D, Polom K, Pascale V, et al. Strong Prognostic Value of Microsatellite Instability in Intestinal Type Non-cardia Gastric Cancer. Ann Surg Oncol 2016; 23: 943-950.
- Shigeyasu K, Nagasaka T, Mori Y, et al. Clinical Significance of MLH1 Methylation and CpG Island Methylator Phenotype as Prognostic Markers in Patients with Gastric Cancer. PLoS One 2015; 10: e0130409.
- Bria E, Pilotto S, Simbolo M, et al. Comprehensive molecular portrait using next generation sequencing of resected intestinal-type gastric cancer patients dichotomized according to prognosis. Sci Rep 2016; 6: 22982.
- Lee HS, Choi SI, Lee HK, et al. Distinct clinical features and outcomes of gastric cancers with microsatellite instability. Mod Pathol 2002; 15: 632-640.
- Zhu L, Li Z, Wang Y, et al. Microsatellite instability and survival in gastric cancer: A systematic review and meta-analysis. Mol Clin Oncol 2015; 3: 699-705.
- Oki E, Kakeji Y, Zhao Y, et al. Chemosensitivity and survival in gastric cancer patients with microsatellite instability. Ann Surg Oncol 2009; 16: 2510-2515.
- An JY, Kim H, Cheong JH, et al. Microsatellite instability in sporadic gastric cancer: its prognostic role and guidance for 5-FU based chemotherapy after R0 resection. Int J Cancer 2012; 131: 505-511.
- Giampieri R, Maccaroni E, Mandolesi A, et al. Mismatch repair deficiency may affect clinical outcome through immune response activation in metastatic gastric cancer patients receiving first-line chemotherapy. Gastric Cancer 2016.
- Bockorny B, Pectasides E. The emerging role of immunotherapy in gastric and esophageal adenocarcinoma. Future Oncol 2016.
- Jou E, Rajdev L. Current and emerging therapies in unresectable and recurrent gastric cancer. World J Gastroenterol 2016; 22: 4812-4823.
- Achille A, Biasi MO, Zamboni G, et al. Cancers of the papilla of vater: mutator phenotype is associated with good prognosis. Clin Cancer Res 1997; 3: 1841-1847.
- Ruemmele P, Dietmaier W, Terracciano L, et al. Histopathologic features and microsatellite instability of cancers of the papilla of vater and their precursor lesions. Am J Surg Pathol 2009; 33: 691-704.
- Agaram NP, Shia J, Tang LH, et al. DNA mismatch repair deficiency in ampullary carcinoma: a morphologic and immunohistochemical study of 54 cases. Am J Clin Pathol 2010; 133: 772-780.
- Achille A, Baron A, Zamboni G, et al. Molecular pathogenesis of sporadic duodenal cancer. Br J Cancer 1998; 77: 760-765.
- Gingras MC, Covington KR, Chang DK et al. Ampullary Cancers Harbor ELF3 Tumor Suppressor Gene Mutations and Exhibit Frequent WNT Dysregulation. Cell Rep 2016; 14: 907-919.
- Yachida S, Wood LD, Suzuki M, et al. Genomic Sequencing Identifies ELF3 as a Driver of Ampullary Carcinoma. Cancer Cell 2016; 29: 229-240.
- Farris AB, 3rd, Demicco EG, Le LP, et al. Clinicopathologic and molecular profiles of microsatellite unstable Barrett Esophagus-associated adenocarcinoma. Am J Surg Pathol 2011; 35: 647-655.
- Baruah A, Buttar N, Chandra R, et al. Translational research on Barrett's esophagus. Ann N Y Acad Sci 2014; 1325: 170-186.
- Tafe LJ. Targeted Next-Generation Sequencing for Hereditary Cancer Syndromes: A Focus on Lynch Syndrome and Associated Endometrial Cancer. J Mol Diagn 2015; 17: 472-482.
- Mills AM, Liou S, Ford JM, et al. Lynch syndrome screening should be considered for all patients with newly diagnosed endometrial cancer. Am J Surg Pathol 2014; 38: 1501-1509.
- Goodfellow PJ, Billingsley CC, Lankes HA, et al. Combined Microsatellite Instability, MLH1 Methylation Analysis, and Immunohistochemistry for Lynch Syndrome Screening in Endometrial Cancers From GOG210: An NRG Oncology and Gynecologic Oncology Group Study. J Clin Oncol 2015; 33: 4301-4308.
- Cohn DE, Frankel WL, Resnick KE, et al. Improved survival with an intact DNA mismatch repair system in endometrial cancer. Obstet Gynecol 2006; 108: 1208-1215.
- Nelson GS, Pink A, Lee S, et al. MMR deficiency is common in high-grade endometrioid carcinomas and is associated with an unfavorable outcome. Gynecol Oncol 2013; 131: 309-314.
- Chu MM, Liu SS, Tam KF, et al. The Significance of Mismatch Repair Deficiency in Young Patients With Endometrial Cancer. Int J Gynecol Pathol 2015; 34: 403-410.
- Diaz-Padilla I, Romero N, Amir E, et al. Mismatch repair status and clinical outcome in endometrial cancer: a systematic review and meta-analysis. Crit Rev Oncol Hematol 2013; 88: 154-167.
- Gargiulo P, Della Pepa C, Berardi S, et al. Tumor genotype and immune microenvironment in POLE-ultramutated and MSI-hypermutated Endometrial Cancers: New candidates for checkpoint blockade immunotherapy? Cancer Treat Rev 2016; 48: 61-68.
- Jensen KC, Mariappan MR, Putcha GV, et al. Microsatellite instability and mismatch repair protein defects in ovarian epithelial neoplasms in patients 50 years of age and younger. Am J Surg Pathol 2008; 32: 1029-1037.
- Segev Y, Pal T, Rosen B, et al. Risk factors for ovarian cancers with and without microsatellite instability. Int J Gynecol Cancer 2014; 24: 664-669.
- Xiao X, Melton DW, Gourley C. Mismatch repair deficiency in ovarian cancer -- molecular characteristics and clinical implications. Gynecol Oncol 2014; 132: 506-512.
- Murphy MA, Wentzensen N. Frequency of mismatch repair deficiency in ovarian cancer: a systematic review This article is a US Government work and, as such, is in the public domain of the United States of America. Int J Cancer 2011; 129: 1914-1922.
- Pal T, Permuth-Wey J, Kumar A, Sellers TA. Systematic review and meta-analysis of ovarian cancers: estimation of microsatellite-high frequency and characterization of mismatch repair deficient tumor histology. Clin Cancer Res 2008; 14: 6847-6854.
- Scartozzi M, De Nictolis M, Galizia E, et al. Loss of hMLH1 expression correlates with improved survival in stage III-IV ovarian cancer patients. Eur J Cancer 2003; 39: 1144-1149.
- Begum FD, Hogdall CK, Kjaer SK, et al. Distribution of microsatellite instability in Danish ovarian tumor patients and the prognostic value in ovarian cancer patients. Oncol Res 2008; 17: 43-49.
- Zhai QJ, Rosen DG, Lu K, et al. Loss of DNA mismatch repair protein hMSH6 in ovarian cancer is histotype-specific. Int J Clin Exp Pathol 2008; 1: 502-509.
- Ercoli A, Ferrandina G, Raspaglio G, et al. hMSH2 and GTBP expression in advanced stage epithelial ovarian cancer. Br J Cancer 1999; 80: 1665-1671.
- Marcelis CL, van der Putten HW, Tops C, et al. Chemotherapy resistant ovarian cancer in carriers of an hMSH2 mutation? Fam Cancer 2001; 1: 107-109.
- Ercoli A, Ferrandina G, Genuardi M, et al. Microsatellite instability is not related to response to cisplatin-based chemotherapy in cervical cancer. Int J Gynecol Cancer 2005; 15: 308-311.
- Roa SJ, Martinez SR, Montenegro S, et al. [Microsatellite instability and human papilloma virus genotypes in preneoplastic and neoplastic uterine cervix lesions]. Rev Med Chil 2007; 135: 37-44.
- Wong YF, Cheung TH, Poon KY, et al. The role of microsatellite instability in cervical intraepithelial neoplasia and squamous cell carcinoma of the cervix. Gynecol Oncol 2003; 89: 434-439.
- Anbazhagan R, Fujii H, Gabrielson E. Microsatellite instability is uncommon in breast cancer. Clin Cancer Res 1999; 5: 839-844.
- Ford JM. Is breast cancer a part of Lynch syndrome? Breast Cancer Res 2012; 14: 110.
- Lotsari JE, Gylling A, Abdel-Rahman WM, et al. Breast carcinoma and Lynch syndrome: molecular analysis of tumors arising in mutation carriers, non-carriers, and sporadic cases. Breast Cancer Res 2012; 14: R90.
- Chiappini F, Gross-Goupil M, Saffroy R, et al. Microsatellite instability mutator phenotype in hepatocellular carcinoma in non-alcoholic and non-virally infected normal livers. Carcinogenesis 2004; 25: 541-547.
- Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol 2006; 21: 15-21.
- Laghi L, Beghelli S, Spinelli A, et al. Irrelevance of microsatellite instability in the epidemiology of sporadic pancreatic ductal adenocarcinoma. PLoS One 2012; 7: e46002.
- Humphris JL, Patch AM, Nones K, et al. Hypermutation in pancreatic cancer. Gastroenterology in press.
- Wilentz RE, Goggins M, Redston M, et al. Genetic, immunohistochemical, and clinical features of medullary carcinoma of the pancreas: A newly described and characterized entity. Am J Pathol 2000; 156: 1641-1651.
- Riazy M, Kalloger SE, Sheffield BS, et al. Mismatch repair status may predict response to adjuvant chemotherapy in resectable pancreatic ductal adenocarcinoma. Mod Pathol 2015; 28: 1383-1389.
- Nakata B, Wang YQ, Yashiro M, et al. Prognostic value of microsatellite instability in resectable pancreatic cancer. Clin Cancer Res 2002; 8: 2536-2540.
- Yamamoto H, Itoh F, Nakamura H, et al. Genetic and clinical features of human pancreatic ductal adenocarcinomas with widespread microsatellite instability. Cancer Res 2001; 61: 3139-3144.
- Cesinaro AM, Ubiali A, Sighinolfi P, et al. Mismatch repair proteins expression and microsatellite instability in skin lesions with sebaceous differentiation: a study in different clinical subgroups with and without extracutaneous cancer. Am J Dermatopathol 2007; 29: 351-358.
- Kruse R, Rutten A, Schweiger N, et al. Frequency of microsatellite instability in unselected sebaceous gland neoplasias and hyperplasias. J Invest Dermatol 2003; 120: 858-864.
- Orta L, Klimstra DS, Qin J, et al. Towards identification of hereditary DNA mismatch repair deficiency: sebaceous neoplasm warrants routine immunohistochemical screening regardless of patient's age or other clinical characteristics. Am J Surg Pathol 2009; 33: 934-944.
- Kubecek O, Trojanova P, Molnarova V, et al. Microsatellite instability as a predictive factor for immunotherapy in malignant melanoma. Med Hypotheses 2016; 93: 74-76.
- Palmieri G, Ascierto PA, Cossu A, et al. Assessment of genetic instability in melanocytic skin lesions through microsatellite analysis of benign naevi, dysplastic naevi, and primary melanomas and their metastases. Melanoma Res 2003; 13: 167-170.
- Subramaniam DS, Liu SV, Giaccone G. Novel approaches in cancer immunotherapy. Discov Med 2016; 21: 267-274.
- Loo K, Daud A. Emerging biomarkers as predictors to anti-PD1/PD-L1 therapies in advanced melanoma. Immunotherapy 2016; 8: 775-784.
- Shen C, Wang X, Tian L, Che G. Microsatellite alteration in multiple primary lung cancer. J Thorac Dis 2014; 6: 1499-1505.
- Ninomiya H, Nomura K, Satoh Y, et al. Genetic instability in lung cancer: concurrent analysis of chromosomal, mini- and microsatellite instability and loss of heterozygosity. Br J Cancer 2006; 94: 1485-1491.
- Kim CH, Yoo CG, Han SK, et al. Genetic instability of microsatellite sequences in non-small cell lung cancers. Lung Cancer 1998; 21: 21-25.
- Warth A, Korner S, Penzel R, et al. Microsatellite instability in pulmonary adenocarcinomas: a comprehensive study of 480 cases. Virchows Arch 2016; 468: 313-319.
- Arai H, Okudela K, Oshiro H, et al. Elevated microsatellite alterations at selected tetra-nucleotide (EMAST) in non-small cell lung cancers--a potential determinant of susceptibility to multiple malignancies. Int J Clin Exp Pathol 2013; 6: 395-410.
- Seng TJ, Currey N, Cooper WA, et al. DLEC1 and MLH1 promoter methylation are associated with poor prognosis in non-small cell lung carcinoma. Br J Cancer 2008; 99: 375-382.
- Eckert A, Kloor M, Giersch A, et al. Microsatellite instability in pediatric and adult high-grade gliomas. Brain Pathol 2007; 17: 146-150.
- Martinez R, Schackert HK, Plaschke J, et al. Molecular mechanisms associated with chromosomal and microsatellite instability in sporadic glioblastoma multiforme. Oncology 2004; 66: 395-403.
- Alonso M, Hamelin R, Kim M, et al. Microsatellite instability occurs in distinct subtypes of pediatric but not adult central nervous system tumors. Cancer Res 2001; 61: 2124-2128.
- Giunti L, Cetica V, Ricci U, et al. Type A microsatellite instability in pediatric gliomas as an indicator of Turcot syndrome. Eur J Hum Genet 2009; 17: 919-927.
- Viana-Pereira M, Lee A, Popov S, et al. Microsatellite instability in pediatric high grade glioma is associated with genomic profile and differential target gene inactivation. PLoS One 2011; 6: e20588.
- Pritchard CC, Morrissey C, Kumar A, et al. Complex MSH2 and MSH6 mutations in hypermutated microsatellite unstable advanced prostate cancer. Nat Commun 2014; 5: 4988.
- Beltran H. DNA mismatch repair in prostate cancer. J Clin Oncol 2013; 31: 1782-1784.
- Haraldsdottir S, Hampel H, Wei L, et al. Prostate cancer incidence in males with Lynch syndrome. Genet Med 2014; 16: 553-557.
- Stoler DL, Datta RV, Charles MA, et al. Genomic instability measurement in the diagnosis of thyroid neoplasms. Head Neck 2002; 24: 290-295.
- Bauer AJ, Cavalli LR, Rone JD, et al. Evaluation of adult papillary thyroid carcinomas by comparative genomic hybridization and microsatellite instability analysis. Cancer Genet Cytogenet 2002; 135: 182-186.
- Mitmaker E, Alvarado C, Begin LR, et al. Microsatellite instability in benign and malignant thyroid neoplasms. J Surg Res 2008; 150: 40-48.
- Santos JC, Bastos AU, Cerutti JM, et al. Correlation of MLH1 and MGMT expression and promoter methylation with genomic instability in patients with thyroid carcinoma. BMC Cancer 2013; 13: 79.
- Onda M, Nakamura I, Suzuki S, et al. Microsatellite instability in thyroid cancer: hot spots, clinicopathological implications, and prognostic significance. Clin Cancer Res 2001; 7: 3444-3449.
- Glavac D, Volavsek M, Potocnik U, et al. Low microsatellite instability and high loss of heterozygosity rates indicate dominant role of the suppressor pathway in squamous cell carcinoma of head and neck and loss of heterozygosity of 11q14.3 correlates with tumor grade. Cancer Genet Cytogenet 2003; 146: 27-32.
- De Schutter H, Spaepen M, Mc Bride WH, et al. The clinical relevance of microsatellite alterations in head and neck squamous cell carcinoma: a critical review. Eur J Hum Genet 2007; 15: 734-741.
- Rubio-Del-Campo A, Salinas-Sanchez AS, Sanchez-Sanchez F, et al. Implications of mismatch repair genes hMLH1 and hMSH2 in patients with sporadic renal cell carcinoma. BJU Int 2008; 102: 504-509.
- Stoehr C, Burger M, Stoehr R, et al. Mismatch repair proteins hMLH1 and hMSH2 are differently expressed in the three main subtypes of sporadic renal cell carcinoma. Pathobiology 2012; 79: 162-168.
- Kawaguchi K, Oda Y, Takahira T, et al. Microsatellite instability and hMLH1 and hMSH2 expression analysis in soft tissue sarcomas. Oncol Rep 2005; 13: 241-246.
- Alldinger I, Schaefer KL, Goedde D, et al. Microsatellite instability in Ewing tumor is not associated with loss of mismatch repair protein expression. J Cancer Res Clin Oncol 2007; 133: 749-759.
- Dolan DE, Gupta S. PD-1 pathway inhibitors: changing the landscape of cancer immunotherapy. Cancer Control 2014; 21: 231-237.
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012; 12: 252-264.
- Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014; 515: 577-581.
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124-128.
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014; 371: 2189-2199.
- Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014; 515: 572-576.
- Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer 2014; 14: 786-800.
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013; 500: 415-421.