Definition of 1p/19q co-deletion
Complete deletion of both the short arm of chromosome 1 (1p) and the long arm of chromosome 19 (19q) (1p/19q co-deletion) is the molecular genetic signature of oligodendrogliomas, a subtype of primary brain tumours accounting for approximately ten to fifteen percent of all diffuse gliomas in adults 1,2. The loss of one hybrid chromosome results in 1p and 19q loss of heterozygosity (LOH) 3. This molecular alteration is the result of an unbalanced whole-arm translocation between chromosomes 1 and 19 3 with the loss of the derivative t(1p;19q), which occurs early in the pathogenesis of oligodendrogliomas. Initially described in 1994 4, the biologic effect of 1p/19q co-deletion remains unclear. 1p/19q co-deletion is a valuable diagnostic, prognostic and predictive biomarker for the management of oligodendroglial tumours.
1p/19q co-deletion as a diagnostic biomarker in glioma
1p/19q co-deletion is a pathognomonic biomarker that defines a distinct glioma entity 5 and is characteristic of oligodendrogliomas 6,7. Virtually, all 1p/19q co-deleted oligodendrogliomas have mutation in isocitrate dehydrogenase 1 (IDH1) at arginine 132 (R132) or the analogous residue arginine 172 in IDH2 (R172) 7,8. Other common molecular alterations co-occuring with 1p/19q co-deletion include mutations in the telomerase reverse transcriptase (TERT) gene promoter, mutations in homolog of Drosophila capicua (CIC) and far upstream element binding protein (FUBP1) 9, and promoter methylation of the methyl-guanine methyl transferase (MGMT) gene 8,7. With very few exceptions, 1p/19q co-deletion is mutually exclusive with TP53 and ATRX mutation, which both characterize glial tumours of astrocytic lineage.
Thereby, assessment of 1p/19q co-deletion, together with IDH mutation status and other molecular markers (e.g. ATRX and TP53 status), can help distinguish oligodendrogliomas which are IDH-mutant and 1p/19q-codeleted, from tumours of astrocytic lineage which are 1p/19q-non co-deleted 6,5.
From a clinical perspective, 1p/19q-co-deleted oligodendrogliomas are predominantly tumours of adulthood, with a peak incidence between the fourth and the sixth decades of life. They tend to present with seizures, and typically involve the frontal lobe with some tumours containing calcifications that are detectable on brain imaging.
1p/19q co-deletion as a prognostic biomarker in glioma
The presence of 1p/19q co-deletion is a strong independent prognostic biomarker associated with improved survival in both diffuse low-grade and anaplastic tumours 3,7,10,11,12.
Among all diffuse gliomas, patients with 1p/19q-co-deletion have the most favourable prognosis 13,7,14. In a large multiomics and clinical retrospective analysis performed by The Cancer Genome Atlas Research Network (TCGA), patients with grades II/III gliomas with an IDH mutation and 1p/19q-co-deletion had a median overall survival of 8.0 years, as compared to 6.3 years in patients with an IDH mutation and no 1p/19q-co-deletion and 1.7 years in patients with wildtype IDH 7.
1p/19q co-deletion as a predictive biomarker in glioma
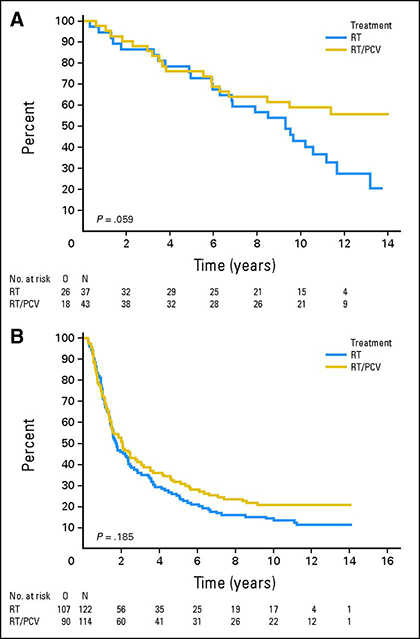
1p/19q-co-deletion has predictive value for response to chemotherapy in anaplastic oligodendrogliomas. Extended follow-up of two large randomised controlled trials that compared procarbazine/lomustine/vincristine (PCV) chemotherapy in combination with radiotherapy to radiotherapy alone demonstrated a survival advantage of first-line chemotherapy in 1p/19q-co-deleted oligodendrogliomas 15,16. Long-term analysis of the EORTC 26951 study that randomised 368 patients to radiotherapy or radiotherapy followed by PCV demonstrated that median overall survival for patients with 1p/19q-co-deleted tumours was 9.3 years for those treated with radiotherapy alone but was not yet reached in those who received radiotherapy plus PCV 15. In a similar analysis of the RTOG 9402 trial in which 289 patients received radiotherapy or PCV followed by radiotherapy, median overall survival doubled from 7.3 to 14.7 years in patients with 1p/19q-co-deleted tumours who received PCV and radiotherapy 16.
Similarly, patients with “high-risk” grade II oligodendroglioma (i.e. patients younger than 40 years of age who had undergone subtotal resection or biopsy or patients who were 40 years of age or older) who received radiotherapy plus PCV had longer overall survival compared to patients treated with radiation therapy alone in the randomised phase III trial RTOG 9802 17.
1p/19q co-deletion testing in glioma
According to the 2016 World Health Organization (WHO) Classification of Tumors of the Central Nervous System, the definitive diagnosis of grade II and grade III (anaplastic) oligodendroglioma requires the demonstration of both an IDH gene family mutation and 1p/19q-co-deletion 6. There are several different methods that have been used to identify patients whose tumours harbour 1p/19q-co-deletion and there is still little clear consensus over how testing should be approached.
Fluorescent in situ hybridisation (FISH) is a reliable, cost-effective method that is capable of detecting the abnormality in minimal amounts of tumour cells on formalin-fixed, paraffin-embedded (FFPE) tissue sections. Dual fluorescent-labelled DNA probes are used to detect 1p and 19q loci within the interphase nuclei of individual glioma cells from FFPE tissue sections transcribed on to unstained slides. Changes in the 1p and 19q probe signals compared with controls are used to determine the presence of 1p/19q-co-deletion. Chromosome 1 and chromosome 19 statuses are assessed, on separate slides, by analyzing the distribution of test and control probes in 50-100 non-overlapping interphase nuclei 18. FISH results may be expressed as a percentage of tumour cells with a deleted signal or as a ratio of test to control probes for each chromosome 18. Pre-specified cut-offs are used to determine whether a chromosome deletion is present or not, and if both 1p and 19q are deleted, 1p/19q co-deletion is reported.
Variability exists between currently available 1p and 19q FISH probes and the chromosome loci they target. Some laboratories use commercially available FISH probes while other centres manufacture their own in-house 18. Furthermore, there is variation in the definitions and cut-offs used to determine chromosome deletion 18. Borderline results that lie near to a pre-defined cut-off for chromosome deletion may require re-testing or clarification with an additional technique. Given the clinical significance of 1p/19q-co-deletion status, it is important that FISH protocols are standardised to ensure that reproducible assessment within and between laboratories. The Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS) have published practical recommendations to assist FISH-based analysis of the 1p/19q status 19.
FISH is unable to differentiate between the whole chromosome arm deletions with centromeric breakpoints characteristic of 1p/19q co-deleted oligodendrogliomas, from smaller focal deletions. This distinction is important given the association of 1p/19q whole-arm co-deletion with improved survival and response to chemotherapy in the oligodendroglial tumour subtype. Array comparative genomic hybridisation (aCGH) and single nucleotide polymorphism (SNP) array are capable of identifying loss of the whole arm of 1p or 19q with higher reliability and may be used in preference to FISH when feasible. However, compared with FISH, these techniques tend to be more labour intensive and require a higher proportion of neoplastic cells 18.
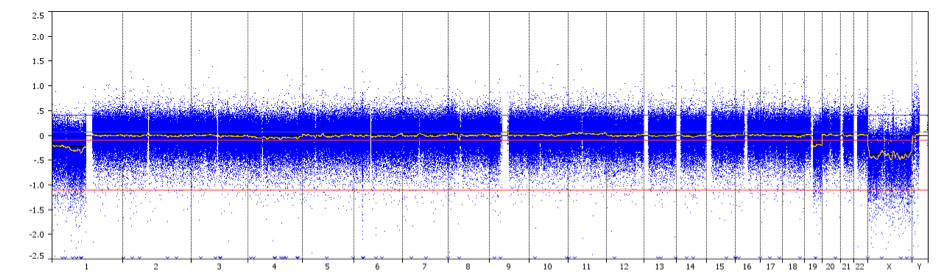
aCGH is a technique to detect genomic copy number variations at high resolution level. DNA – extracted from a test sample and control sample – are labelled using different fluorescent dyes, mixed together and hybridized to several thousand probes. Digital imaging systems are used to quantify the fluorescence intensities of the labelled DNA probes that have hybridized to each probe. The fluorescence ratio of the test and control hybridization signals is determined at different positions along the genome, providing information on the relative copy number of sequences in the test genome as compared to a normal genome. This method allows simultaneous detection of chromosome aneuploidies, deletions, duplications, and/or amplifications of any locus plotted on an array.
Finally, polymerase chain reaction-based microsatellite analysis – that allows detection of LOH at selected loci, and next-generation sequencing – NGS-based methods can also be used to assess 1p/19q status.
Patient selection
European guidelines recommend that1p/19q-co-deletion should be evaluated to support a diagnosis of oligodendroglioma and to predict the chemosensitivity and prognosis of these patients 5,20. On the basis of the EORTC 26951 and RTOG 9402 trial findings, patients with 1p/19q-co-deleted anaplastic oligodendroglial tumours should not be treated with radiotherapy alone, but instead receive early alkylating chemotherapy with radiotherapy 20. A similar approach should be considered in a subset of patients with grade II oligodendroglioma.
When medical treatment needs to be initiated, the magnitude of treatment benefit from combined radiotherapy plus PCV is substantial, but the toxic effects are greater in patients receiving the combination. Clinical trials are ongoing to evaluate the value of upfront chemotherapy alone or combined radiotherapy plus temozolomide as compared to standard radiotherapy plus PCV.
Key References
- Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol 2014; 16(7):896-913.
- Ibdaih A, Marie Y, Pierron G, et al. Two types of chromosome 1p losses with opposite significance in gliomas. Ann Neurol 2005; 58(3):483-7.
- Jenkins R, Blair H, Ballman K, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 2006; 66: 9852–61.
- Reifenberger J, Reifenberger G, Liu L, et al. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol 1994; 145:1175–90.
- Stupp R, Brada M, van den Bent M, et al. High-grade glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2014;25 (Suppl 3): iii93-101.
- Louis D, Perry A, Reifenberger G. et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathologica 2016; 131: 803-20.
- Cancer Genome Atlas Research Network, Brat D, Verhaak R, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 2015; 372: 2481-98.
- Sanson M, Marie Y, Paris S, et al. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J Clin Oncol 2009; 27: 4150–54.
- Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 2011; 333(6048): 1453-5.
- Zhao J, Ma W, Zhao H. Loss of heterozygosity 1p/19q and survival in glioma: a meta-analysis. Neuro-Oncology 2014; 16: 103–112.
- van den Bent M, Carpentier A, Brandes A, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol 2006; 24: 2715–22
- Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol 2006; 24: 2707-14.
- Wiestler B, Capper D, Sill M, et al. Integrated DNA methylation and copy‑number profiling identify three clinically and biologically relevant groups of anaplastic glioma. Acta Neuropathologica 2014; 128:561-71.
- Ceccarelli M, Barthel FP, Malta TM, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016; 164(3): 550-63.
- van den Bent M, Brandes A, Taphoorn M, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol 2013; 31:344–50.
- Cairncross G, Wang M, Shaw E, et al. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol 2013; 31: 337–43.
- Buckner JC, Shaw EG, Pugh SL, et al. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N Engl J Med 2016; 374: 1344-1355.
- Woehrer A, Sander P, Baberler C, et al. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice – a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS). Clinical Neuropathology 2011; 30: 47-55.
- Pinkham M, Telford N, Whitfield G, et al. FISHing Tips: What Every Clinician Should Know About 1p19q Analysis in Gliomas Using Fluorescence in situ Hybridisation. Clinical Oncology 2015; 27: 445-53.
- Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol 2014; 15: 395-403.