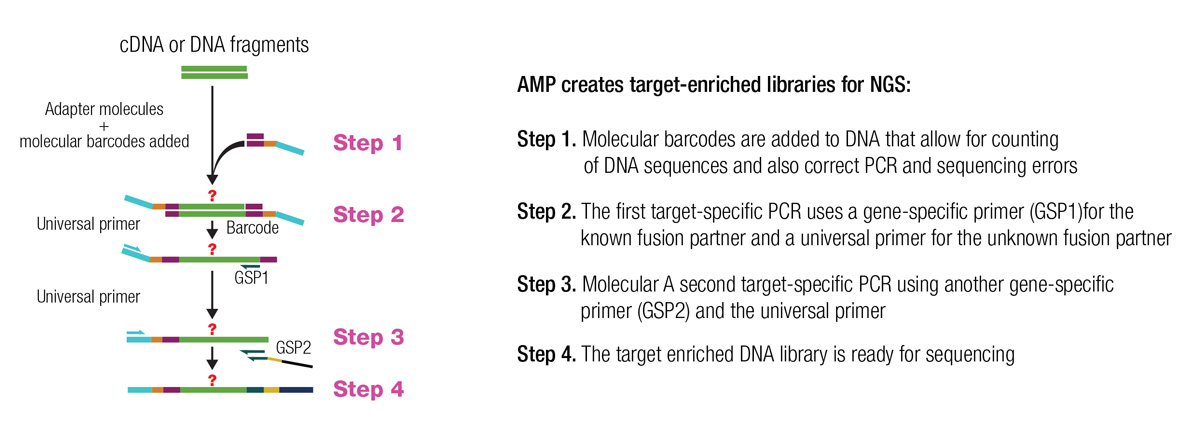
AMP is a form of targeted NGS that allows detection of fusions and identification of partner genes from low nucleic acid input sourced from formalin-fixed paraffin-embedded specimens [1]. After RNA extraction, cDNA is made. Double-stranded cDNA undergoes end repair, adenylation and ligation with a universal half-functional adapter [1]. The library is rendered functional after two subsequent rounds of nested low-cycle PCR, which are the core steps for target enrichment. The second PCR step uses nested primers with 5′ tags for a common sequencing adapter. The resulting target amplicons are functionalised for clonal amplification (for example, emulsion PCR or bridge PCR) and sequencing. Nontarget fragments remain half-functional and do not need to be removed from the library. Libraries are quantitated and processed for Illumina or Ion Torrent sequencing. The following figure shows the basic approach utilised in AMP.
AMP for the Detection of Gene Rearrangements
The advantages and disadvantages of AMP are summarised below.
Advantages and Disadvantages of AMP for Testing for NTRK Gene Fusions
Advantages |
Disadvantages |
|---|---|
|
|
References
- Zheng Z, Liebers M, Zhelyazkova B et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 2014; 20: 1479-1484.
- Lam SW, Cleton-Jansen AM, Cleven AHG et al. Molecular Analysis of Gene Fusions in Bone and Soft Tissue Tumors by Anchored Multiplex PCR-Based Targeted Next-Generation Sequencing. J Mol Diagn 2018; 20: 653-663.