Prof. Le Tourneau explains what we need in terms of multigene sequencing
Definition
Multigene sequencing is the use of next-generation sequencing (NGS) with the ability to assess multiple gene molecular alterations in the same assay. From this breakthrough technology has emerged the term “precision medicine” in oncology, which is according to the National Cancer Institute “a form of medicine that uses information about a person’s genes, proteins, and environment to prevent, diagnose, and treat disease”. DNA gene sequencing allows seeking for mutations or gene copy number alterations, whilst RNA sequencing allows seeking for gene fusions, validate somatic mutations found in DNA as well as altered gene expression and splicing. Different types of sequencing exist, including whole genome sequencing (WGS), whole exome sequencing (WES) and targeted sequencing. WGS and WES require constitutional DNA in addition to tumour DNA for somatic variant calling. WGS and WES also allow the evaluation of mutational and neoantigen loads that might be of interest to predict response to some immunotherapeutic agents, as well as genomic instability for DNA repair inhibitors (1, 2).
Predictive Value
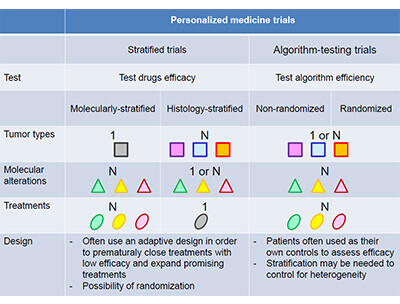
The predictive value of multigene sequencing cannot be interpreted the same way than for a single molecular alteration. The key question behind the predictive value of multigene sequencing is whether the use of multigene sequencing improves patient outcomes (clinical utility). The predictive value of multigene sequencing is only assessed in trials using multigene sequencing to guide therapy. These trials are dichotomized into stratified trials that include basket trials (histologic stratification) and umbrella trials (molecular stratification), and algorithm-testing trials that mix molecular alterations, drugs and often tumour types (3).
The stratified trials are designed in such a way that they can conclude whether a specific drug has any activity in a molecularly- and histologically-characterised subgroup of patients. Therefore, these trials do not assess the predictive value of multigene sequencing but of any of the genes tested individually. In contrast, algorithm-testing trials are not powered to evaluate whether a specific drug has any activity in a specific subgroup of patients, and can only eventually indicate whether the use of multigene sequencing to guide therapy according to a specific treatment-algorithm improves patient outcomes. The treatment algorithm encompasses several features including the technology used, the thresholds used to claim a molecular alteration is a driver, the matches between molecular alterations and drugs, and prioritization of molecular alterations (4).
Several non-randomised algorithm-testing trials have suggested that multigene sequencing might improve patient outcomes (5, 6, 7, 8). However, the only randomised trial (SHIVA) did not confirm these results (9). In summary, there is no level 1 evidence to date demonstrating the predictive value of multigene sequencing.
Prognostic Value
It is well established that some molecular alterations have a prognostic value, such as EGFR mutations in lung cancer. The question of whether the molecular alterations detected when using multigene sequencing have a prognostic value is key. Indeed, some non-randomised studies retrospectively comparing the outcome of patients treated with matched therapy based on the identification of a druggable molecular alteration with the outcome of patients who were not treated with matched therapy (one of the reasons being the lack of identification of a druggable molecular alteration) reported a statistically significant better outcome in the matched group (6, 7). Due to the lack of randomisation, nobody can tell whether the observed difference is due the predictive value of multigene sequencing or its prognostic value. The overall survival analysis of SHIVA suggests that patients with a druggable molecular alteration have indeed a better prognosis than those who have not (HR=0.85), although it did not reach statistical significance (10). In summary, we cannot exclude that multigene sequencing has a prognostic value. The prognostic value of multigene sequencing obviously depends on the molecular alterations that are being assessed.
Testing Recommendations
Multigene sequencing is today used in precision medicine trials or screening programmes in comprehensive cancer centers worldwide aiming to orient patients towards appropriate early phase clinical trials. A key issue for multigene sequencing is the choice of the design taking into account the relevant list of genes to be tested, the desire to detect in addition to mutations, gene copy number alterations, the heterogeneity of the tumour sample with a normal and tumour components and the type of tissue (DNA from frozen samples or formalin-fixed, paraffin-embedded [FFPE] samples or circulating tumour DNA [ctDNA]). For targeted sequencing, hybridization-based and amplicon-based capture methods differ in their ability to uniformly capture and sequence targeted regions and identify mutations (11). Amplicon-based targeted sequencing is adapted for simplified wet labs workflow and utilizes smaller DNA inputs. It is consequently well adapted to FFPE DNA and ctDNA, while hybridization capture based NGS is more performant in terms of sequencing complexity and coverage uniformity (11). The design of the targeted panel should take into account 1) the type of gene to insure a complete coverage of all exons of tumour suppressor genes while coverage can be limited to hotspots for known oncogenes, and 2) the choice of primers covering extremities of genes to evaluate the size of alteration when copy number variations are evaluated. It is important to differentiate large versus focal alterations for gene specificities (4). In addition, the depth of coverage is crucial to identify mutations from minor sub-clones or to exclude false positive variants (4), especially in the context of heterogeneous tissue. WES, a hybridization capture based sequencing, encompassing all exons of our 20000 genes, is today well adapted to clinical practice in light of the decreasing costs. However, the bioinformatics analyses need to be adapted to the clinics. A simple recommendation would be to focus on the 400 known driver genes for concise molecular report. Comprehensive genomic profiling is necessary to evaluate tumour mutational burden.
Ensuring Quality and Timely Testing Results
Multigene gene sequencing for clinical trials and routine diagnostics implies a standardized process from sample collection, assessment of tissue quality, nucleic acid extraction, and wet labs assays to bioinformatics pipelines. Quality control checkpoints at each step are therefore a prerequisite and are implemented in FDA or ISO certified labs. Bioinformatics scripts able to identify a risk of contamination need to be implemented and to report directly to the wet labs. The depth of coverage is proportional to the sensitivity of detecting mutations in a heterogeneous tissue samples and thus needs to be adjusted for samples with limited tumour content. The accuracy of variant detection is strongly influenced by the quality of the computational pipeline to avoid false positive variants’ detection. Guidelines are available for the bioinformatics analyses and interpretation of results but these guidelines need to be adjusted in the context of the rapid evolution of sequencing techniques (12). Expert biological interpretation remains crucial following bioinformatics to validate driver molecular alterations (classification of pathogen versus benign variants). In some cases, alternative techniques such as immunohistochemistry, FISH or Sanger sequencing can be required to discriminate potential false positive alterations. The integration of the different types of data including clinical and molecular and the flow of information between the different stakeholders has to be meticulously orchestrated to ensure timely results adapted to the clinic.
Patient Selection
There is no level 1 evidence that multigene sequencing improves patient outcome to date. Now, multigene sequencing is a useful tool for guiding patients to clinical trials enriched based on a specific molecular alteration. Multigene sequencing avoids preforming multiple sequential single tests with the following advantages:
- spare tissue samples
- avoid losing time for the patients and
- be able to direct the patient right to the most appropriate clinical trial.
In the context clinical trial orientation based on molecular alterations, multigene sequencing is recommended for patients who have a priori no contra-indication for an inclusion in clinical trials, such as a good performance status and adequate haematological, renal and hepatic functions. Multigene sequencing should be encouraged if clinical trials are eventually available. In summary, patient selection should be based on patient characteristics and the availability of clinical trials.
References
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9(1):34.
- Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16(2):110-120.
- Le Tourneau C, Kamal M, Alt M, et al. The spectrum of clinical trials aiming at personalizing medicine. Chin Clin Oncol 2014;3:13.
- Le Tourneau C, Kamal M, Tsimberidou AM, et al. Treatment Algorithms Based on Tumor Molecular Profiling: The Essence of Precision Medicine Trials. J Natl Cancer Inst 2015;108(4).
- von Hoff DD, Stephenson JJ, Rosen P, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol 2010;28(33):4877-4883.
- Tsimberidou AM, Iskander NG, Hong DS, et al. Personalized medicine in a phase I clinical trials program: The M. D. Anderson Cancer Center Initiative. Clin Cancer Res 2012;18:6373–6383.
- Tsimberidou AM, Wen S, Hong DS, et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: validation and landmark analyses. Clin Cancer Res 2014;20:4827–4836.
- Massard C, Michiels S, Ferte C, et al. High-Throughput Genomics and Clinical Outcome in Hard-to-Treat Advanced Cancers: Results of the MOSCATO 01 Trial. Cancer Discov 2017;7(6):586-595.
- Le Tourneau C, Delord JP, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 2015;16(13): 1324-1334.
- Dureau S, Ricci F, Alt M, et al. SHIVA - Randomized phase II trial comparing molecularly targeted therapy based on tumor molecular profiling versus conventional therapy in patients with refractory cancer: Overall survival (OS) analysis. J Clin Oncol 35, 2017 (suppl; abstr 11515).
- Samorodnitsky E, Jewell BM, Hagopian R, et al. Evaluation of Hybridization Capture Versus Amplicon-Based Methods for Whole-Exome Sequencing. Hum Mutat 2015;36(9):903-914.
- Li MM, Datto M, Duncavage EJ, et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19(1):4-23.